Help
Input types for enzymes/proteins and metabolites
Enzymes
The input for an enzyme must be a string containing the enzyme's amino acid sequence. The model currently does not handle missing information, such as the '*' character, and including it will prevent the model from proceeding.
Metabolites
There are three valid input types for the metabolite: SMILES string, KEGG Compound ID, and InChI string
InChI string
InChI strings are textual representations of chemical structures. Every InChI string is a unique identifier and contains detailed information about the structure of a small molecule. For more details on InChI, see this page from IUPAC.
KEGG Compound ID
The KEGG Compound database contains identifiers for many small molecules and drugs. A KEGG Compound ID starts with a "C" or "D" followed by a five-digit number. For more information see the KEGG homepage.
SMILES
Simplified Molecular Input Line Entry Specification (SMILES) allows to represent the structure of a molecule using ASCII strings. You can get the SMILES for a molecule e.g. by searching for the molecules name in PubChem. Since SMILES representations are not unique for all molecules, we recommend to use InChI string or KEGG Compound IDs instead, if possible.
Single Input File
Case study: ESP (Enzyme-Substrate Pair Prediction) step by step
-
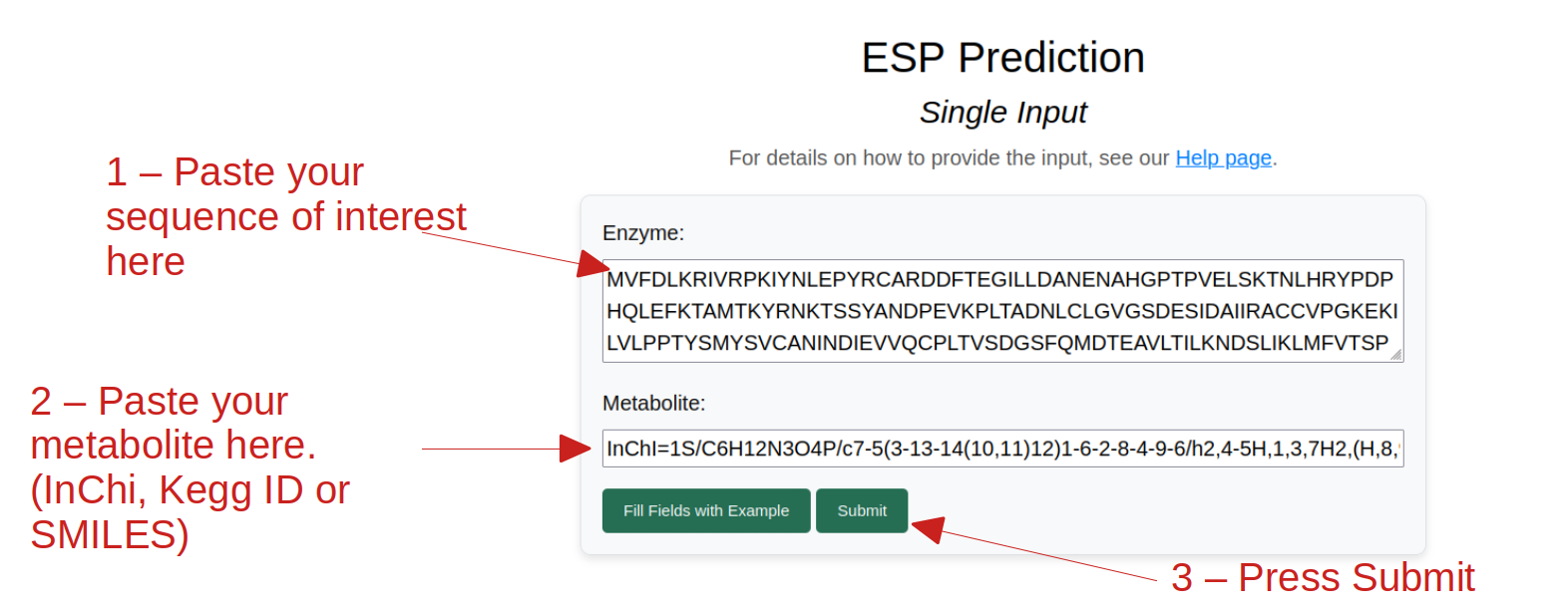
If you are interested in predicting the interaction between a single enzyme and a single metabolite, you can use the single input option. You can enter the enzyme sequence and the metabolite in the respective fields and click on the "Predict" button. The metabolite can be entered as a SMILES string, KEGG Compound ID, or InChI string. To see an example, you can click on "Fill Fields with Example".
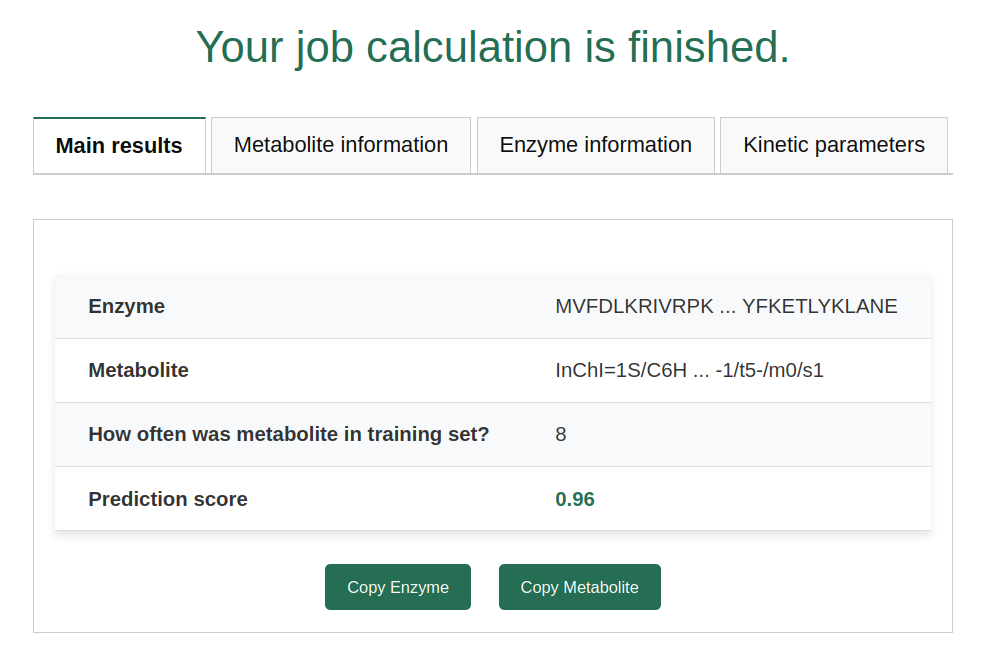
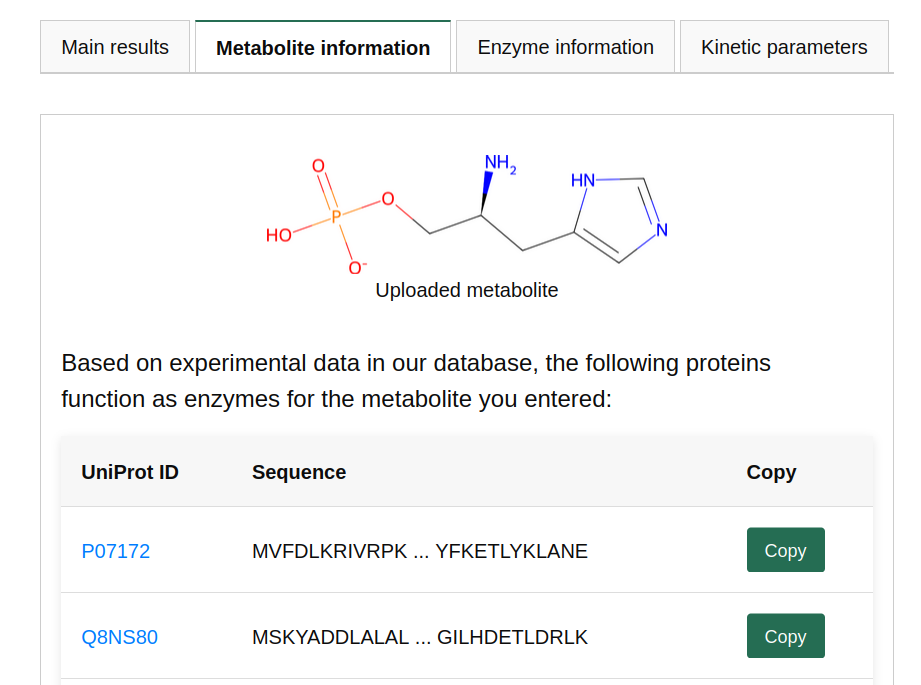
Prediction scores close to 0.5 (i.e., scores in the range of 0.3 to 0.7) should be considered with caution. The prediction model is unsure which class to predict in these cases. The model also returns how often the metabolite was present in the training set. In this case, the metabolite was present in the training set 8 times. We have shown that the prediction performance of our model is low when applied to metabolites that were not present in our training set. Therefore, we check whether every uploaded metabolite was part of our training set.
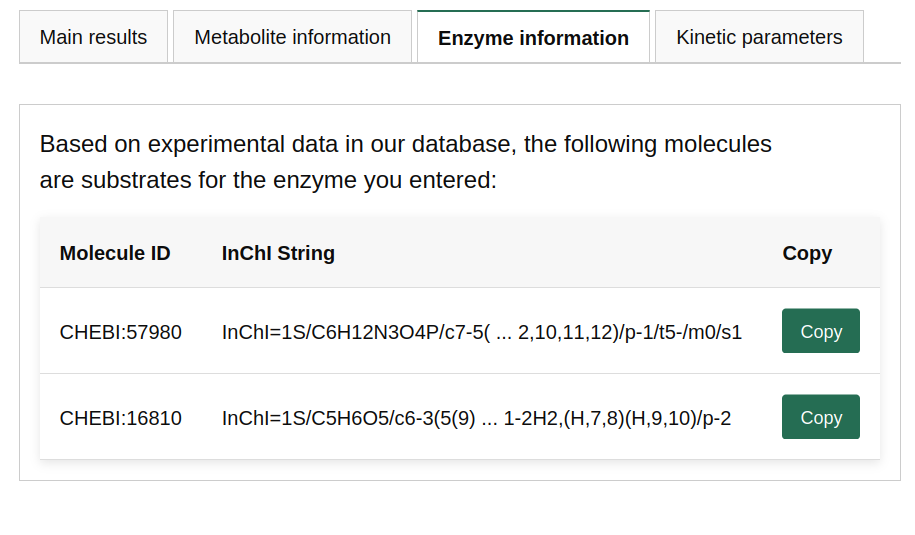
TurNuP (kcat prediction)
Predicting the turnover number kcat of an enzyme-reaction pair works similarly to the case study above. The main difference is that, to optimize the prediction, the model requires information about the catalyzed reaction. This is done by providing several substrates and products, using semicolons as separators.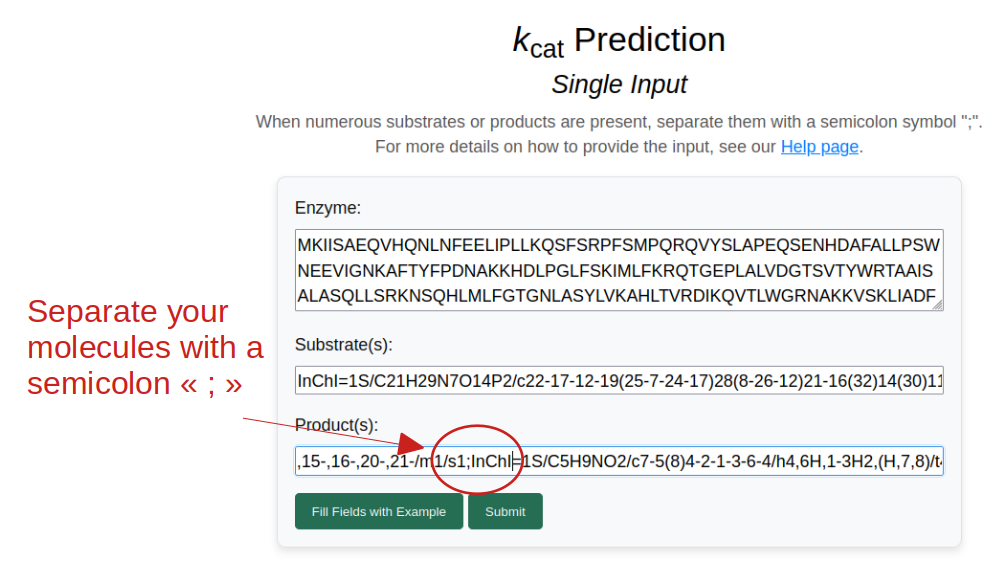
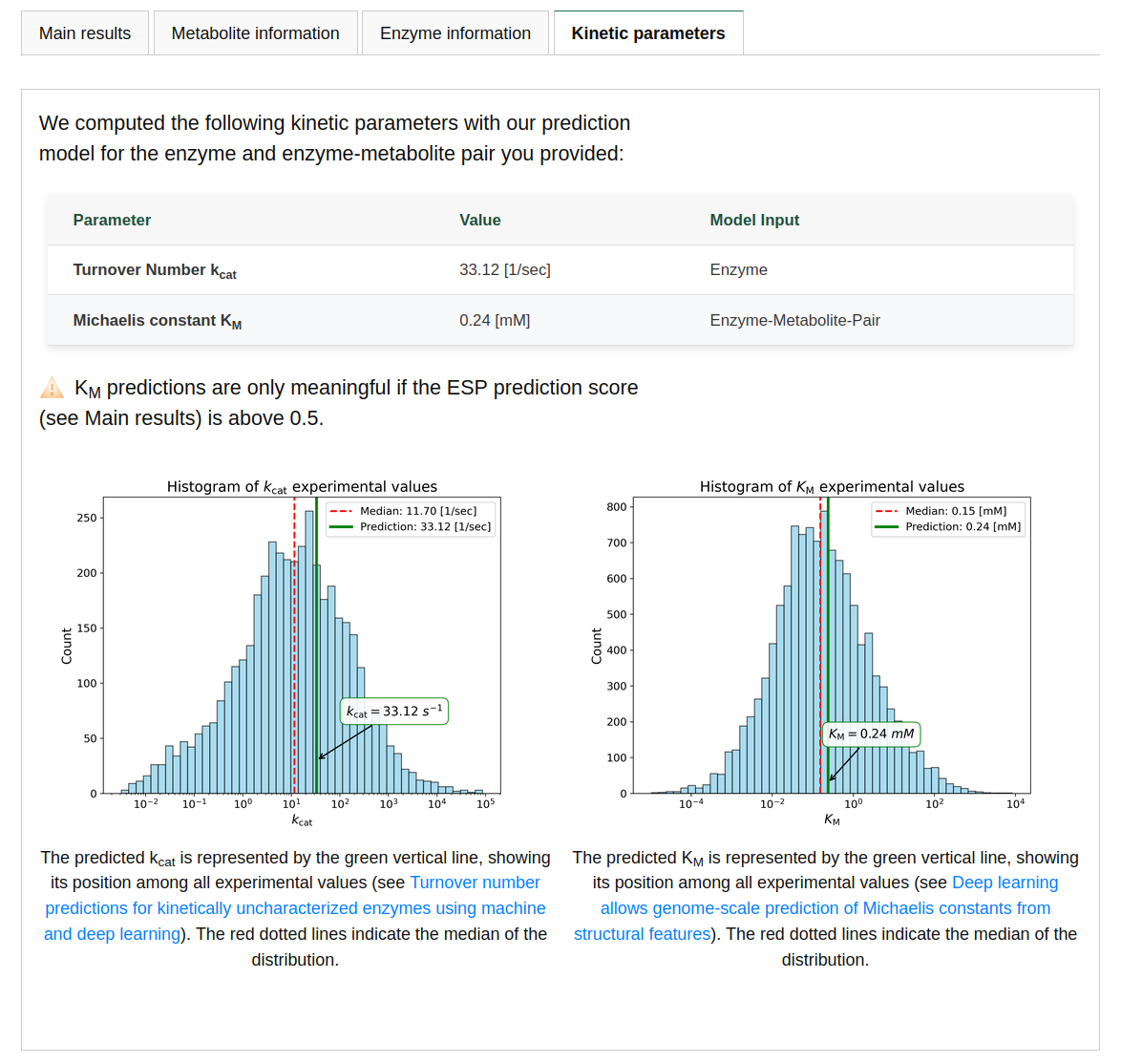
KM prediction
Predicting the Michaelis constant KM of an enzyme-substrate pair works similarly to the case study above. On average, predictions for new enzyme-substrate deviate from the true KM value by a factor of 4.1 (see our manuscript for more details). On an independent test set, the model achieves a coefficient of determination R2R2 of 0.53. If similar enzymes or similar molecules were part of our training set, the model's accuracy increases. We also provide additional information and predictions, such as other sequences catalyzing the same substrate, other molecules catalyzed by the same enzyme, and, finally, an estimate of the turnover number kcatt for the enzyme-substrate pair. Here, kcat is estimated solely based on the amino acid sequence and might therefore be less accurate than when reaction information is provided. You can use the "Example" button to fill the fields with an example and examine the different outputs.SPOT (Transporter - Substrate Pair prediction)
Predicting the interaction between a transporter and a substrate works similarly to the case study above. The prediction score is a value between 0 and 1. Scores close to 1 mean that the model predicts that the molecule is likely a substrate for the given transporter, whereas scores close to 0 mean that the model predicts that the molecule is likely not a substrate for the transporter. Prediction scores close to 0.5 (i.e. scores in the range of 0.4 to 0.6) should be considered with caution. The prediction model is unsure which class it should predict in these cases. We have shown that the prediction performance of our model is better when substrates are present in our training set. Therefore, we check for every uploaded molecule if it was part of our training set. We also provide additional information, such as other transporter of the same substrate and other molecules that are substrates of the same transporter. You can use the "Example" button to fill the fields with an example and examine the different outputs.Multiple Input file
Switch from CLS to XLSX format
Starting from December 9, 2024, we have switched from using CLS files to XLSX files specifically for kcat predictions. XLSX files can be created using spreadsheet programs like Microsoft Excel or Google Sheets, or through pandas library in python. For more details on how to create an XLSX file, you can refer to Microsoft Excel or Google Sheets.
How should your file look like?
Your file format depends on the model you are using. Attention: InChI strings can contain commas (","), so be sure to properly structure your data in the required format.
You can download a sample file for each model below.
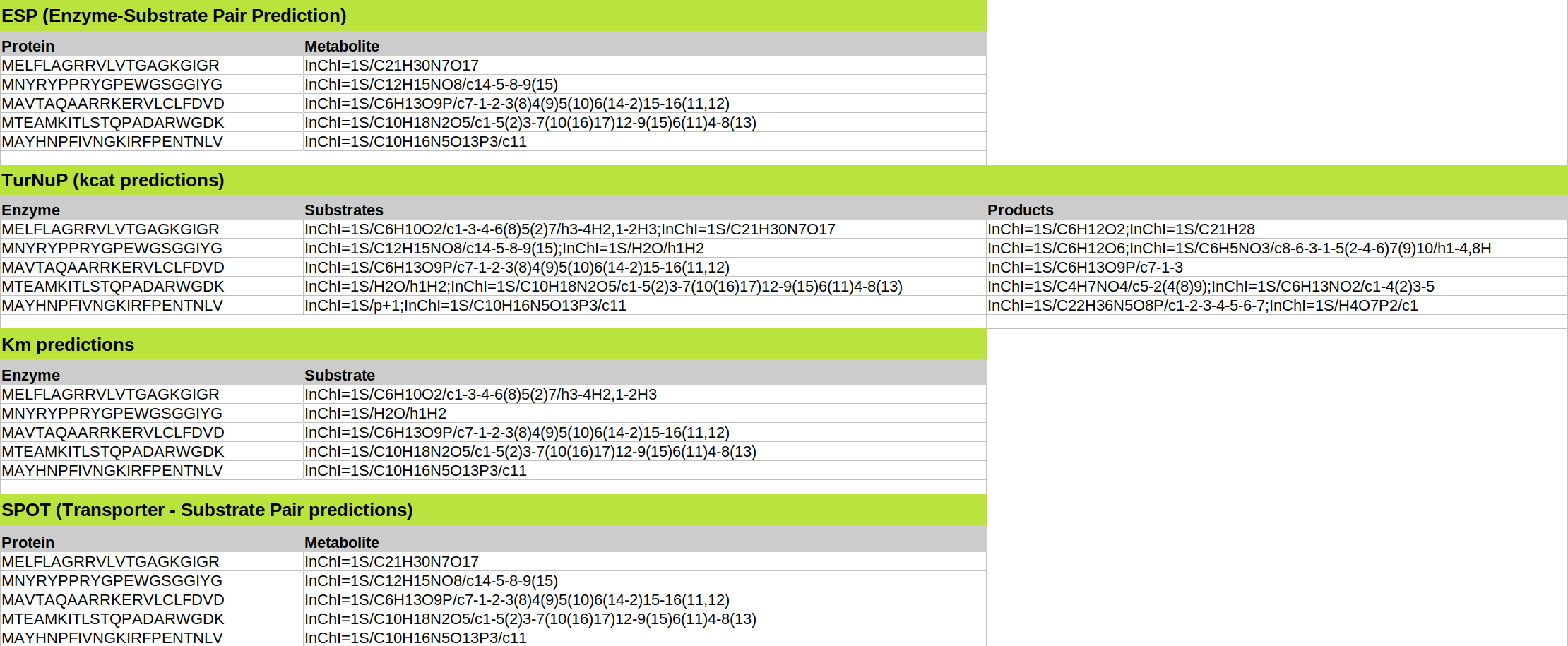
Example of multiple inputs with a file. The enzyme-substrate pairs and metabolites displayed here are not real.
-
Enzyme-Substrate Pair Prediction:
Your file must be in XLSX format and contain exactly two columns, one called "Protein" and one called "Metabolite". Each row should contain one enzyme and one metabolite in the format described above. The upper limit of accepted enzyme-metabolite pairs is 500. You can download a sample file here. We have shown that the prediction performance of our model is low when it is applied to metabolites which were not present in our training set. Therefore, we check for every uploaded metabolite if it was part of our training set. We return this information in the column "metabolite".
-
kcat prediction:
Your file must be in XLSX format and contain three columns, titled "Enzyme", "Substrates", and "Products". Each row should contain one enzyme-reaction pair in the format described above. For both columns Substrates and Products, metabolites should be separated by a semicolon ";". The upper limit of accepted enzyme-reaction pairs is 500. You can download a sample XLSX file here.
KM prediction:Your file must be in XLSX format and contain exactly two columns, one called "Enzyme" and one called "Substrate". Each row should contain one enzyme and one metabolite in the described format. The upper limit of accepted enzyme-metabolite pairs is 500. You can download a sample file here.
SPOT:Your file must be in XLSX format and contain exactly two columns, one called "Protein" and one called "Metabolite". Each row should contain one transporter and one molecule in the format described above. The upper limit of accepted transporter-molecule pairs is 1000. You can download a sample file here.
© 2019 All Rights Reserved By Free Html Templates